Annotations and Scout Data
Spots of interest can be annotated by attaching labels to them. Labeling is a flexible and powerful tool to manage the annotations and additional data retrieved by DECODON Scouts.
Annotations: Labels
Managing master gels with spot identifications is easy – a single click will transfer labels from one gel to another. You can change label colors and formats according to your preferences.
For labelled gel coordinates you can easily export a picklist. If you like, translate intermediate spot annotations into real protein names by importing a list of identified proteins.
In combination with DECODON Scouts, you get access to additional information (e.g. from the UniProt Database) about each protein, including external links, sequence, and biochemical parameters.
Just like any other data generated by Delta2D, label data is saved in XML files that can easily be processed by other applications.
Sophisticated Label Management
Label Selection
Want to select a number of labels and copy this selection to another image? No problem, label selection is now available.
For selected labels the same operations are available as for single labels:
- Delete
- Adjust
- Move
- Copy
Label Grouping
Having identified spots you learn that some spots contain the same protein? Avoid multiple labes with the same text by grouping them - you just keep one label pointing to the different spots.
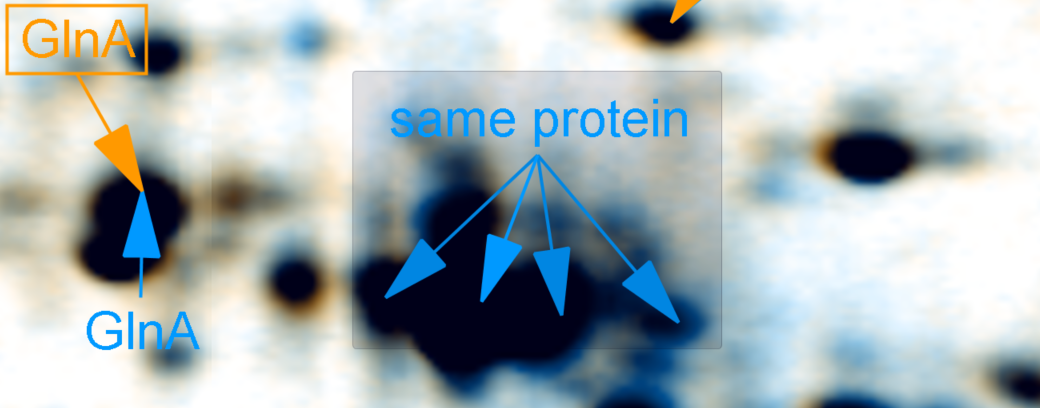
Label Translation
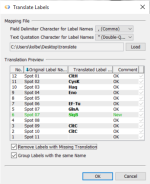
No need to copy and paste protein identifications into the label fields to annotate spots according to the MS results. Just import a translation list containing old and new label texts. the list can even include multiple identifications - you will receive multiple labels for the same spot then.
If the list contains the same identifications for more than one spot you can let Delta2D automatically group them so that you see just one label pointing to every spot containing this protein.
DECODON Scouts

DECODON Scouts automate work that would have to be a tedious job: Submit a query for a certain protein name in the web database of your preference (e.g. UniProt) and copy and paste the interesting attributes from the resulting page the database is presenting.
A certain Scout automatically searches in the respective database, takes the Protein's properties and saves them along with the label in your analysis project. You can then just open a Label report to get a list including the complete available data.
Automatic label color coding illustrates numerical protein properties using different colors. Data like pI, MW, protein size and so on can be visualized by continous color gradients. This, for example, makes it easy to find proteins with unusual pI or MW caused by posttranslational modifications.
